Introduction
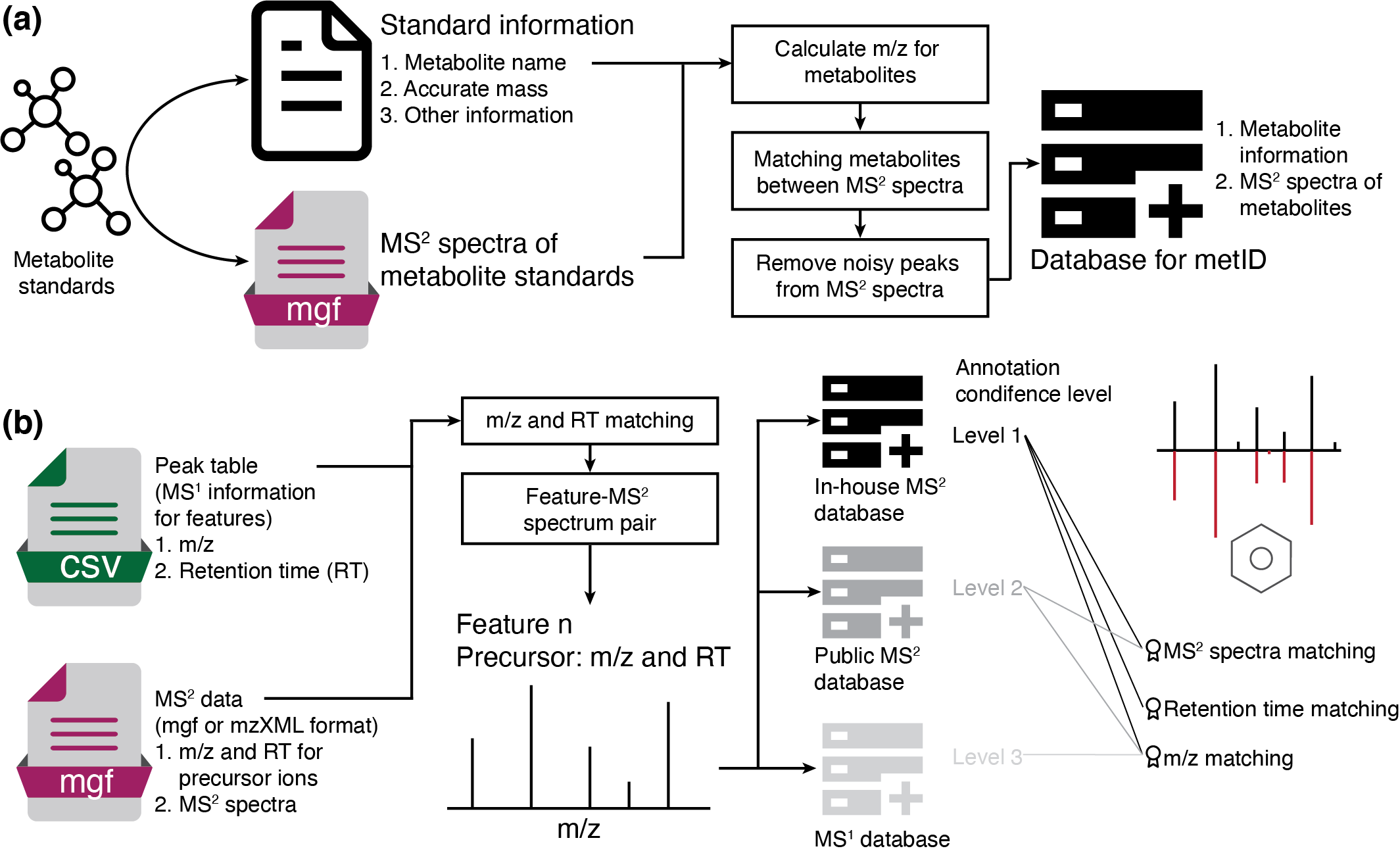
In tidyMass, metid package is used for metabolite identification based
on in-house database and public database based on accurate mass (m/z),
retention time (RT) and/or MS2 spectra.
Installation
You can install metid from GitLab.
if(!require(remotes)){
install.packages("remotes")
}
remotes::install_gitlab("tidymass/metid")
or GitHub
remotes::install_github("tidymass/metid")
or tidymass.org
source("https://www.tidymass.org/tidymass-packages/install_tidymass.txt")
install_tidymass(from = "tidymass.org", which_package = "metid")
Citation
If you use metid in your publications, please cite this paper:
-
Xiaotao Shen, Si Wu, Liang Liang, Songjie Chen, Kevin Contrepois, Zheng-Jiang Zhu*, Michael Snyder* (Corresponding Author). metID: A R package for automatable compound annotation for LC−MS-based data. Bioinformatics, btab583, https://doi.org/10.1093/bioinformatics/btab583
-
Shen, X., Yan, H., Wang, C. et al. TidyMass an object-oriented reproducible analysis framework for LC–MS data. Nat Commun 13, 4365 (2022). Weblink
Thanks very much!