Install tidymass
Xiaotao Shen (https://www.shenxt.info/)
Created on 2021-12-04 and updated on 2022-03-09
tidymass_install.RmdUpdate R version
tidymass require R version > 4.1. You can check your R version in your console:
versionIf your R version is < 4.1, please download and install the latest version of R, and then restart your R.
Uninstall tidyTools and metID
If you have installed tinyTools or metID, please uninstall them.
tinyTools has been renamed as tinytools and metID has been renamed as metid. So please uninstall them before you install tidymass. And don’t use them any more.
- detach them by:
- Remove them by:
remove.packages(pkgs = "tinyTools")
remove.packages(pkgs = "metID")- Restart your R session.
If there are errors, ignore them.
Install tidymass from GitLab
Update remotes
You can use the remotes package to install tidymass.
Please update remotes first and then restart your r session.
install.packages("remotes")Install tidymass
Install tidymass by:
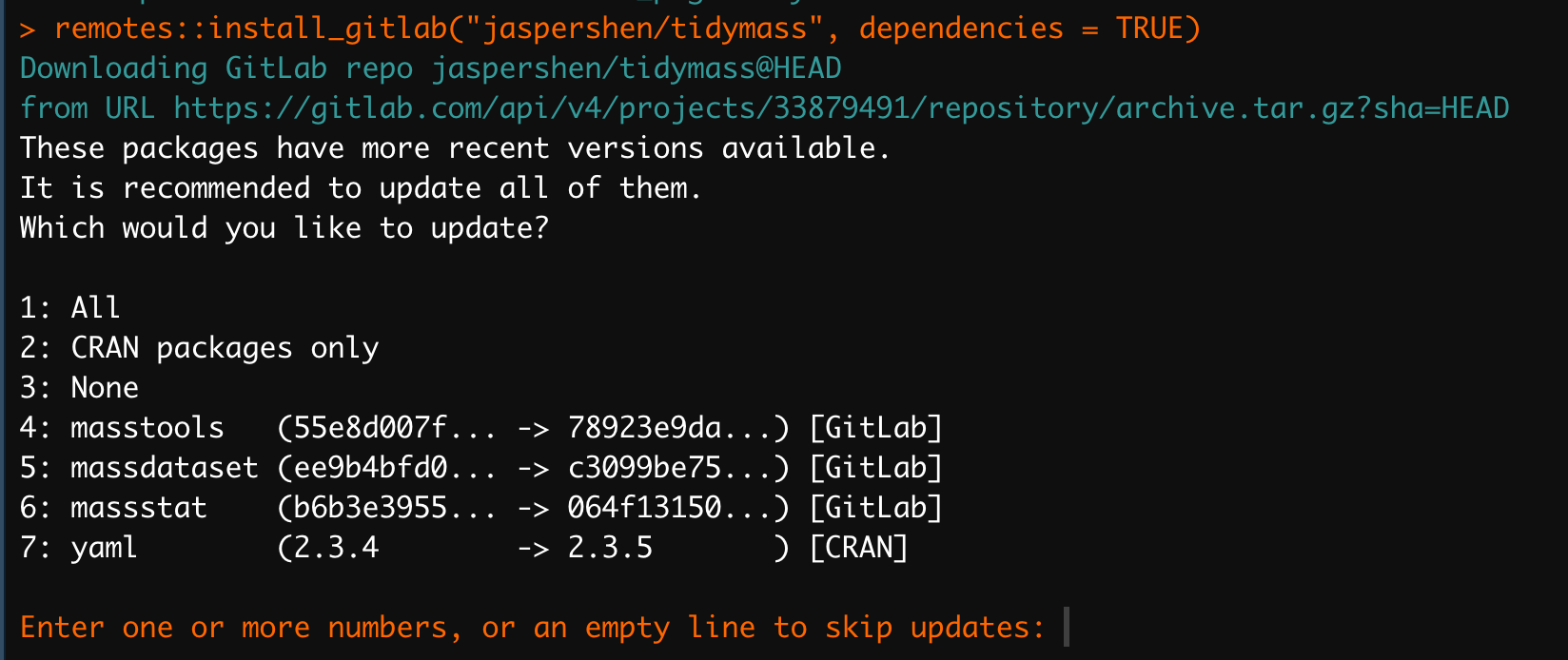
remotes::install_gitlab("jaspershen/tidymass", dependencies = TRUE)During installing, it may ask you several times: “Would you like to uodate some pacakges?” Just Enter the
EnterorRetrunkey to skip updates.
Install tidymass packages one by one
If you use the Windows OS and the dependencies can not be installed, try to install all the tidymass packages one by one.
remotes::install_gitlab("jaspershen/masstools", dependencies = TRUE)
remotes::install_gitlab("jaspershen/massdataset", dependencies = TRUE)
remotes::install_gitlab("jaspershen/massqc", dependencies = TRUE)
remotes::install_gitlab("jaspershen/massprocesser", dependencies = TRUE)
remotes::install_gitlab("jaspershen/masscleaner", dependencies = TRUE)
remotes::install_gitlab("jaspershen/massstat", dependencies = TRUE)
remotes::install_gitlab("jaspershen/metid", dependencies = TRUE)
remotes::install_gitlab("jaspershen/metpath", dependencies = TRUE)
Install tidymass from GitHub
Install tidymass
Then install tidymass:
remotes::install_github("tidymass/tidymass", dependencies = TRUE)During the installation, it will ask if you want to update some packages for few times, just enter
EnterorReurnkey to skip it.
If there is a error like below:
Error: Failed to install 'tidymass' from GitHub: HTTP error 403. API rate limit exceeded for 171.66.10.237. (But here's the good news: Authenticated requests get a higher rate limit. Check out the documentation for more details.)Try to resolve it by:
- In you R console, type this code:
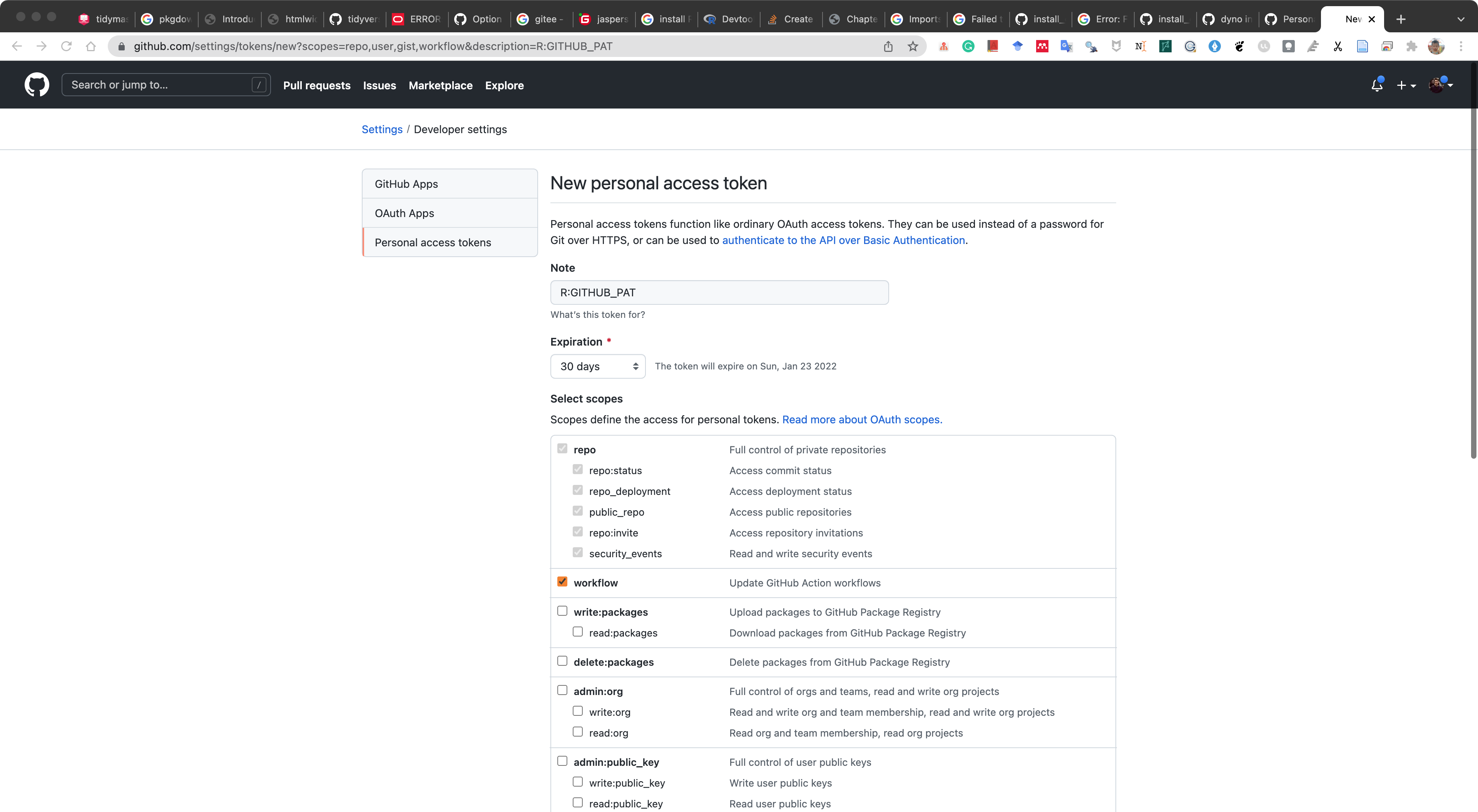
usethis::create_github_token()It will open a page in browser, and create a “New personal access token” and copy it.
- Then type this code:
usethis::edit_r_environ()and then add one line like below:
GITHUB_PAT=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The
GITHUB_PATshould be yours that is created in step 1.
And then restart R session and try again.
Install tidymass packages one by one
If you use the Windows OS and the dependencies can not be installed, try to install all the tidymass packages one by one.
remotes::install_github("tidymass/masstools", dependencies = TRUE)
remotes::install_github("tidymass/massdataset", dependencies = TRUE)
remotes::install_github("tidymass/massqc", dependencies = TRUE)
remotes::install_github("tidymass/massprocesser", dependencies = TRUE)
remotes::install_github("tidymass/masscleaner", dependencies = TRUE)
remotes::install_github("tidymass/massstat", dependencies = TRUE)
remotes::install_github("tidymass/metid", dependencies = TRUE)
remotes::install_github("tidymass/metpath", dependencies = TRUE)
Install tidymass from Gitee
If you can’t install pacakgs from GitHub and GitLab, please try install packags from Gitee.
Install tidymass
Then install tidymass:
remotes::install_git(url = "https://gitee.com/jaspershen/tidymass", dependencies = TRUE)Install tidymass packages one by one.
If you use the Windows OS and the dependencies can not be installed, try to install all the tidymass packages one by one.
remotes::install_git(url = "https://gitee.com/jaspershen/masstools", dependencies = TRUE)
remotes::install_git(url = "https://gitee.com/jaspershen/massdataset", dependencies = TRUE)
remotes::install_git(url = "https://gitee.com/jaspershen/massqc", dependencies = TRUE)
remotes::install_git(url = "https://gitee.com/jaspershen/massprocesser", dependencies = TRUE)
remotes::install_git(url = "https://gitee.com/jaspershen/masscleaner", dependencies = TRUE)
remotes::install_git(url = "https://gitee.com/jaspershen/massstat", dependencies = TRUE)
remotes::install_git(url = "https://gitee.com/jaspershen/metid", dependencies = TRUE)
remotes::install_git(url = "https://gitee.com/jaspershen/metpath", dependencies = TRUE)Frequently Asked Questions
- Can not install dependent packages
rasterandCario
raster is a package in CRAN, try to install it first and then install tidymass.
install.packages("raster")Cario is a package in CRAN, try to install it first and then install tidymass.
install.packages("Cairo")- Error when install
massprocesser
The error is:
Error: .onLoad failed in loadNamespace() for 'affy', details:
call: assign(".affyInternalEnv", .affyInternalEnv, envir = topenv(parent.frame()))
error: cannot add binding of '.affyInternalEnv' to the base environment
Execution halted
ERROR: lazy loading failed for package 'massprocesser'
* removing 'D:/R_packages/R4.0/library/massprocesser'Just try to restart R and install affy.
BiocManager::install("affy")Session information
sessionInfo()
#> R version 4.1.2 (2021-11-01)
#> Platform: x86_64-apple-darwin17.0 (64-bit)
#> Running under: macOS Big Sur 10.16
#>
#> Matrix products: default
#> BLAS: /Library/Frameworks/R.framework/Versions/4.1/Resources/lib/libRblas.0.dylib
#> LAPACK: /Library/Frameworks/R.framework/Versions/4.1/Resources/lib/libRlapack.dylib
#>
#> locale:
#> [1] en_US.UTF-8/en_US.UTF-8/en_US.UTF-8/C/en_US.UTF-8/en_US.UTF-8
#>
#> attached base packages:
#> [1] stats4 stats graphics grDevices utils datasets methods
#> [8] base
#>
#> other attached packages:
#> [1] dplyr_1.0.8 metid_1.2.2 metpath_0.99.2
#> [4] massstat_0.99.6 ggfortify_0.4.14 ggplot2_3.3.5
#> [7] massqc_0.99.3 masscleaner_0.99.3 xcms_3.16.1
#> [10] MSnbase_2.20.4 ProtGenerics_1.26.0 S4Vectors_0.32.3
#> [13] mzR_2.28.0 Rcpp_1.0.8 Biobase_2.54.0
#> [16] BiocGenerics_0.40.0 BiocParallel_1.28.3 massprocesser_0.99.3
#> [19] magrittr_2.0.2 masstools_0.99.3 massdataset_0.99.9
#> [22] tidymass_0.99.4
#>
#> loaded via a namespace (and not attached):
#> [1] ragg_1.2.1 tidyr_1.2.0
#> [3] missForest_1.4 knitr_1.37
#> [5] DelayedArray_0.20.0 data.table_1.14.2
#> [7] rpart_4.1.16 KEGGREST_1.34.0
#> [9] RCurl_1.98-1.5 doParallel_1.0.17
#> [11] generics_0.1.2 snow_0.4-4
#> [13] leaflet_2.1.0 preprocessCore_1.56.0
#> [15] mixOmics_6.18.1 RANN_2.6.1
#> [17] proxy_0.4-26 future_1.23.0
#> [19] tzdb_0.2.0 xml2_1.3.3
#> [21] lubridate_1.8.0 ggsci_2.9
#> [23] SummarizedExperiment_1.24.0 assertthat_0.2.1
#> [25] tidyverse_1.3.1 viridis_0.6.2
#> [27] xfun_0.29 hms_1.1.1
#> [29] jquerylib_0.1.4 evaluate_0.15
#> [31] DEoptimR_1.0-10 fansi_1.0.2
#> [33] dbplyr_2.1.1 readxl_1.3.1
#> [35] igraph_1.2.11 DBI_1.1.2
#> [37] htmlwidgets_1.5.4 MsFeatures_1.3.0
#> [39] rARPACK_0.11-0 purrr_0.3.4
#> [41] ellipsis_0.3.2 RSpectra_0.16-0
#> [43] crosstalk_1.2.0 backports_1.4.1
#> [45] ggcorrplot_0.1.3 MatrixGenerics_1.6.0
#> [47] vctrs_0.3.8 remotes_2.4.2
#> [49] cachem_1.0.6 withr_2.4.3
#> [51] ggforce_0.3.3 itertools_0.1-3
#> [53] robustbase_0.93-9 checkmate_2.0.0
#> [55] cluster_2.1.2 lazyeval_0.2.2
#> [57] crayon_1.5.0 ellipse_0.4.2
#> [59] pkgconfig_2.0.3 tweenr_1.0.2
#> [61] GenomeInfoDb_1.30.0 nnet_7.3-17
#> [63] rlang_1.0.1 globals_0.14.0
#> [65] lifecycle_1.0.1 affyio_1.64.0
#> [67] extrafontdb_1.0 fastDummies_1.6.3
#> [69] MassSpecWavelet_1.60.0 modelr_0.1.8
#> [71] cellranger_1.1.0 randomForest_4.7-1
#> [73] rprojroot_2.0.2 polyclip_1.10-0
#> [75] matrixStats_0.61.0 Matrix_1.4-0
#> [77] reprex_2.0.1 base64enc_0.1-3
#> [79] GlobalOptions_0.1.2 png_0.1-7
#> [81] viridisLite_0.4.0 rjson_0.2.21
#> [83] clisymbols_1.2.0 bitops_1.0-7
#> [85] pander_0.6.4 Biostrings_2.62.0
#> [87] shape_1.4.6 stringr_1.4.0
#> [89] parallelly_1.30.0 robust_0.7-0
#> [91] readr_2.1.2 jpeg_0.1-9
#> [93] gridGraphics_0.5-1 scales_1.1.1
#> [95] memoise_2.0.1 plyr_1.8.6
#> [97] zlibbioc_1.40.0 compiler_4.1.2
#> [99] RColorBrewer_1.1-2 pcaMethods_1.86.0
#> [101] clue_0.3-60 rrcov_1.6-2
#> [103] cli_3.2.0 affy_1.72.0
#> [105] XVector_0.34.0 listenv_0.8.0
#> [107] patchwork_1.1.1 pbapply_1.5-0
#> [109] htmlTable_2.4.0 Formula_1.2-4
#> [111] MASS_7.3-55 tidyselect_1.1.1
#> [113] vsn_3.62.0 stringi_1.7.6
#> [115] forcats_0.5.1 textshaping_0.3.6
#> [117] yaml_2.3.4 latticeExtra_0.6-29
#> [119] MALDIquant_1.21 ggrepel_0.9.1
#> [121] grid_4.1.2 sass_0.4.0
#> [123] tools_4.1.2 parallel_4.1.2
#> [125] circlize_0.4.14 rstudioapi_0.13
#> [127] MsCoreUtils_1.6.0 foreach_1.5.2
#> [129] foreign_0.8-82 gridExtra_2.3
#> [131] farver_2.1.0 mzID_1.32.0
#> [133] ggraph_2.0.5 rvcheck_0.2.1
#> [135] digest_0.6.29 BiocManager_1.30.16
#> [137] GenomicRanges_1.46.1 broom_0.7.12
#> [139] ncdf4_1.19 httr_1.4.2
#> [141] ComplexHeatmap_2.10.0 colorspace_2.0-2
#> [143] rvest_1.0.2 XML_3.99-0.8
#> [145] fs_1.5.2 IRanges_2.28.0
#> [147] splines_4.1.2 yulab.utils_0.0.4
#> [149] graphlayouts_0.8.0 pkgdown_2.0.2
#> [151] ggplotify_0.1.0 plotly_4.10.0
#> [153] systemfonts_1.0.3 fit.models_0.64
#> [155] jsonlite_1.7.3 tidygraph_1.2.0
#> [157] corpcor_1.6.10 R6_2.5.1
#> [159] Hmisc_4.6-0 pillar_1.7.0
#> [161] htmltools_0.5.2 glue_1.6.1
#> [163] fastmap_1.1.0 class_7.3-20
#> [165] codetools_0.2-18 pcaPP_1.9-74
#> [167] mvtnorm_1.1-3 furrr_0.2.3
#> [169] utf8_1.2.2 lattice_0.20-45
#> [171] bslib_0.3.1 tibble_3.1.6
#> [173] zip_2.2.0 openxlsx_4.2.5
#> [175] Rttf2pt1_1.3.9 survival_3.2-13
#> [177] limma_3.50.0 rmarkdown_2.11
#> [179] desc_1.4.0 munsell_0.5.0
#> [181] e1071_1.7-9 GetoptLong_1.0.5
#> [183] GenomeInfoDbData_1.2.7 iterators_1.0.14
#> [185] impute_1.68.0 haven_2.4.3
#> [187] reshape2_1.4.4 gtable_0.3.0
#> [189] extrafont_0.17