Data preparation
We just use the dataset which are from previous step.
library(tidymass)
#> Registered S3 method overwritten by 'Hmisc':
#> method from
#> vcov.default fit.models
#> ── Attaching packages ──────────────────────────────────────── tidymass 1.0.9 ──
#> ✔ massdataset 1.0.34 ✔ metid 1.2.34
#> ✔ massprocesser 1.0.10 ✔ masstools 1.0.13
#> ✔ masscleaner 1.0.12 ✔ dplyr 1.1.4
#> ✔ massqc 1.0.7 ✔ ggplot2 3.5.1
#> ✔ massstat 1.0.6 ✔ magrittr 2.0.3
#> ✔ metpath 1.0.8
load("data_cleaning/POS/object_pos2")
load("data_cleaning/NEG/object_neg2")
Add MS2 spectra data to mass_dataset class
Download the MS2 data here.
Uncompress it.
Positive mode
object_pos2 <-
mutate_ms2(
object = object_pos2,
column = "rp",
polarity = "positive",
ms1.ms2.match.mz.tol = 15,
ms1.ms2.match.rt.tol = 30,
path = "mgf_ms2_data/POS"
)
#> Reading mgf data...
#> Reading mgf data...
#> Reading mgf data...
#> Reading mgf data...
#> 1042 out of 5101 variable have MS2 spectra.
#> Selecting the most intense MS2 spectrum for each peak...
object_pos2
#> --------------------
#> massdataset version: 0.99.8
#> --------------------
#> 1.expression_data:[ 5101 x 259 data.frame]
#> 2.sample_info:[ 259 x 6 data.frame]
#> 259 samples:sample_06 sample_103 sample_11 ... sample_QC_38 sample_QC_39
#> 3.variable_info:[ 5101 x 6 data.frame]
#> 5101 variables:M70T53_POS M70T527_POS M71T775_POS ... M836T610_POS M836T759_POS
#> 4.sample_info_note:[ 6 x 2 data.frame]
#> 5.variable_info_note:[ 6 x 2 data.frame]
#> 6.ms2_data:[ 1042 variables x 951 MS2 spectra]
#> --------------------
#> Processing information
#> 9 processings in total
#> Latest 3 processings show
#> normalize_data ----------
#> Package Function.used Time
#> 1 masscleaner normalize_data() 2024-09-25 21:03:38
#> integrate_data ----------
#> Package Function.used Time
#> 1 masscleaner integrate_data() 2024-09-25 21:03:38
#> mutate_ms2 ----------
#> Package Function.used Time
#> 1 massdataset mutate_ms2() 2024-09-25 21:09:39
extract_ms2_data(object_pos2)
#> $`QEP_SGA_QC_posi_ms2_ce25_01.mgf;QEP_SGA_QC_posi_ms2_ce25_02.mgf;QEP_SGA_QC_posi_ms2_ce50_01.mgf;QEP_SGA_QC_posi_ms2_ce50_02.mgf`
#> --------------------
#> column: rp
#> polarity: positive
#> mz_tol: 15
#> rt_tol (second): 30
#> --------------------
#> 1042 variables:
#> M71T775_POS M72T53_POS M83T50_POS M84T57_POS M85T54_POS...
#> 951 MS2 spectra.
#> mz70.981170654297rt775.4286 mz72.081642150879rt53.6528862 mz82.945625305176rt49.238013 mz84.045127868652rt59.6895132 mz85.029016959043rt53.0835648...
Negative mode
object_neg2 <-
mutate_ms2(
object = object_neg2,
column = "rp",
polarity = "negative",
ms1.ms2.match.mz.tol = 15,
ms1.ms2.match.rt.tol = 30,
path = "mgf_ms2_data/NEG"
)
#> Reading mgf data...
#> Reading mgf data...
#> Reading mgf data...
#> Reading mgf data...
#> 1092 out of 4104 variable have MS2 spectra.
#> Selecting the most intense MS2 spectrum for each peak...
object_neg2
#> --------------------
#> massdataset version: 0.99.8
#> --------------------
#> 1.expression_data:[ 4104 x 259 data.frame]
#> 2.sample_info:[ 259 x 6 data.frame]
#> 259 samples:sample_06 sample_103 sample_11 ... sample_QC_38 sample_QC_39
#> 3.variable_info:[ 4104 x 6 data.frame]
#> 4104 variables:M70T712_NEG M70T587_NEG M71T587_NEG ... M884T57_NEG M899T56_NEG
#> 4.sample_info_note:[ 6 x 2 data.frame]
#> 5.variable_info_note:[ 6 x 2 data.frame]
#> 6.ms2_data:[ 1092 variables x 988 MS2 spectra]
#> --------------------
#> Processing information
#> 9 processings in total
#> Latest 3 processings show
#> normalize_data ----------
#> Package Function.used Time
#> 1 masscleaner normalize_data() 2024-09-25 21:03:40
#> integrate_data ----------
#> Package Function.used Time
#> 1 masscleaner integrate_data() 2024-09-25 21:03:40
#> mutate_ms2 ----------
#> Package Function.used Time
#> 1 massdataset mutate_ms2() 2024-09-25 21:09:50
extract_ms2_data(object_neg2)
#> $`QEP_SGA_QC_neg_ms2_ce25_01.mgf;QEP_SGA_QC_neg_ms2_ce25_02.mgf;QEP_SGA_QC_neg_ms2_ce50_01.mgf;QEP_SGA_QC_neg_ms2_ce50_02.mgf`
#> --------------------
#> column: rp
#> polarity: negative
#> mz_tol: 15
#> rt_tol (second): 30
#> --------------------
#> 1092 variables:
#> M71T51_NEG M73T74_NEG M75T52_NEG M80T299_NEG M80T232_NEG...
#> 988 MS2 spectra.
#> mz71.012359619141rt52.3270968 mz73.02799987793rt74.779476 mz75.007308959961rt24.1557228 mz79.955728954783rt301.268466 mz79.955834350356rt235.127328...
Metabolite annotation
Metabolite annotation is based on the metid package.
Download database
We need to download MS2 database from metid website.
Here we download the Michael Snyder RPLC databases, Orbitrap database and MoNA database. And place them in a new folder named as metabolite_annotation.
Positive mode
Annotate features using snyder_database_rplc0.0.3.
load("metabolite_annotation/snyder_database_rplc0.0.3.rda")
snyder_database_rplc0.0.3
#> -----------Base information------------
#> Version:0.0.2
#> Source:MS
#> Link:http://snyderlab.stanford.edu/
#> Creater:Xiaotao Shen(shenxt1990@163.com)
#> With RT information
#> -----------Spectral information------------
#> 14 items of metabolite information:
#> Lab.ID; Compound.name; mz; RT; CAS.ID; HMDB.ID; KEGG.ID; Formula; mz.pos; mz.neg (top10)
#> 917 metabolites in total.
#> 356 metabolites with spectra in positive mode.
#> 534 metabolites with spectra in negative mode.
#> Collision energy in positive mode (number:):
#> Total number:2
#> NCE25; NCE50
#> Collision energy in negative mode:
#> Total number:2
#> NCE25; NCE50
object_pos2 <-
annotate_metabolites_mass_dataset(object = object_pos2,
ms1.match.ppm = 15,
rt.match.tol = 30,
polarity = "positive",
database = snyder_database_rplc0.0.3)
Annotate features using orbitrap_database0.0.3.
load("metabolite_annotation/orbitrap_database0.0.3.rda")
orbitrap_database0.0.3
#> -----------Base information------------
#> Version:0.0.1
#> Source:NIST
#> Link:https://www.nist.gov/
#> Creater:Xiaotao Shen(shenxt1990@163.com)
#> Without RT informtaion
#> -----------Spectral information------------
#> 8 items of metabolite information:
#> Lab.ID; Compound.name; mz; RT; CAS.ID; HMDB.ID; KEGG.ID; Formula
#> 8360 metabolites in total.
#> 7103 metabolites with spectra in positive mode.
#> 3311 metabolites with spectra in negative mode.
#> Collision energy in positive mode (number:):
#> Total number:12
#> 10; 15; 45; 55; 5; 20; 30; 35; 40; 25
#> Collision energy in negative mode:
#> Total number:12
#> 10; 25; 5; 15; 20; 30; 50; 35; 40; 45
object_pos2 <-
annotate_metabolites_mass_dataset(object = object_pos2,
ms1.match.ppm = 15,
polarity = "positive",
database = orbitrap_database0.0.3)
Annotate features using mona_database0.0.3.
load("metabolite_annotation/mona_database0.0.3.rda")
orbitrap_database0.0.3
#> -----------Base information------------
#> Version:0.0.1
#> Source:NIST
#> Link:https://www.nist.gov/
#> Creater:Xiaotao Shen(shenxt1990@163.com)
#> Without RT informtaion
#> -----------Spectral information------------
#> 8 items of metabolite information:
#> Lab.ID; Compound.name; mz; RT; CAS.ID; HMDB.ID; KEGG.ID; Formula
#> 8360 metabolites in total.
#> 7103 metabolites with spectra in positive mode.
#> 3311 metabolites with spectra in negative mode.
#> Collision energy in positive mode (number:):
#> Total number:12
#> 10; 15; 45; 55; 5; 20; 30; 35; 40; 25
#> Collision energy in negative mode:
#> Total number:12
#> 10; 25; 5; 15; 20; 30; 50; 35; 40; 45
object_pos2 <-
annotate_metabolites_mass_dataset(object = object_pos2,
ms1.match.ppm = 15,
polarity = "positive",
database = mona_database0.0.3)
Negative mode
Annotate features using snyder_database_rplc0.0.3.
object_neg2 <-
annotate_metabolites_mass_dataset(object = object_neg2,
ms1.match.ppm = 15,
rt.match.tol = 30,
polarity = "negative",
database = snyder_database_rplc0.0.3)
Annotate features using orbitrap_database0.0.3.
object_neg2 <-
annotate_metabolites_mass_dataset(object = object_neg2,
ms1.match.ppm = 15,
polarity = "negative",
database = orbitrap_database0.0.3)
Annotate features using mona_database0.0.3.
object_neg2 <-
annotate_metabolites_mass_dataset(object = object_neg2,
ms1.match.ppm = 15,
polarity = "negative",
database = mona_database0.0.3)
Annotation result
The annotation result will assign into mass_dataset class as the annotation_table slot.
head(extract_annotation_table(object = object_pos2))
#> variable_id
#> 1 M100T160_POS
#> 2 M104T51_POS
#> 3 M113T187_POS
#> 4 M113T81_POS
#> 5 M113T81_POS
#> 6 M113T81_POS
#> ms2_files_id
#> 1 QEP_SGA_QC_posi_ms2_ce25_01.mgf;QEP_SGA_QC_posi_ms2_ce25_02.mgf;QEP_SGA_QC_posi_ms2_ce50_01.mgf;QEP_SGA_QC_posi_ms2_ce50_02.mgf
#> 2 QEP_SGA_QC_posi_ms2_ce25_01.mgf;QEP_SGA_QC_posi_ms2_ce25_02.mgf;QEP_SGA_QC_posi_ms2_ce50_01.mgf;QEP_SGA_QC_posi_ms2_ce50_02.mgf
#> 3 QEP_SGA_QC_posi_ms2_ce25_01.mgf;QEP_SGA_QC_posi_ms2_ce25_02.mgf;QEP_SGA_QC_posi_ms2_ce50_01.mgf;QEP_SGA_QC_posi_ms2_ce50_02.mgf
#> 4 QEP_SGA_QC_posi_ms2_ce25_01.mgf;QEP_SGA_QC_posi_ms2_ce25_02.mgf;QEP_SGA_QC_posi_ms2_ce50_01.mgf;QEP_SGA_QC_posi_ms2_ce50_02.mgf
#> 5 QEP_SGA_QC_posi_ms2_ce25_01.mgf;QEP_SGA_QC_posi_ms2_ce25_02.mgf;QEP_SGA_QC_posi_ms2_ce50_01.mgf;QEP_SGA_QC_posi_ms2_ce50_02.mgf
#> 6 QEP_SGA_QC_posi_ms2_ce25_01.mgf;QEP_SGA_QC_posi_ms2_ce25_02.mgf;QEP_SGA_QC_posi_ms2_ce50_01.mgf;QEP_SGA_QC_posi_ms2_ce50_02.mgf
#> ms2_spectrum_id Compound.name CAS.ID HMDB.ID
#> 1 mz100.076248168945rt158.377638 N-Methyl-2-pyrrolidone 872-50-4 <NA>
#> 2 mz104.107467651367rt49.510314 5-Amino-1-pentanol 2508-29-4 <NA>
#> 3 mz113.060150146484rt188.406384 1,4-Cyclohexanedione <NA> <NA>
#> 4 mz113.035087585449rt77.20827 Uracil 66-22-8 HMDB00300
#> 5 mz113.035087585449rt77.20827 Maleic hydrazide 123-33-1 <NA>
#> 6 mz113.035087585449rt77.20827 Uracil 66-22-8 <NA>
#> KEGG.ID Lab.ID Adduct mz.error mz.match.score RT.error RT.match.score
#> 1 C11118 MONA_11509 (M+H)+ 1.335652 0.9960435 NA NA
#> 2 <NA> NO07238 (M+H)+ 1.169128 0.9969671 NA NA
#> 3 <NA> MONA_14519 (M+H)+ 1.051626 0.9975454 NA NA
#> 4 C00106 NO07292 (M+H)+ 1.218964 0.9967035 NA NA
#> 5 <NA> NO07300 (M+H)+ 1.218964 0.9967035 NA NA
#> 6 C00106 MONA_2762 (M+H)+ 1.275544 0.9963909 NA NA
#> CE SS Total.score Database Level
#> 1 35 (nominal) 0.6871252 0.7442896 MoNA_0.0.1 2
#> 2 5 0.5971697 0.6906242 NIST_0.0.1 2
#> 3 HCD (NCE 20-30-40%) 0.5401414 0.6566000 MoNA_0.0.1 2
#> 4 10 0.6484578 0.7213092 NIST_0.0.1 2
#> 5 20 0.5486434 0.6614205 NIST_0.0.1 2
#> 6 Ramp 5-60 0.5404285 0.6563874 MoNA_0.0.1 2
variable_info_pos <-
extract_variable_info(object = object_pos2)
head(variable_info_pos)
#> variable_id mz rt na_freq na_freq.1 na_freq.2 Compound.name
#> 1 M70T53_POS 70.06596 52.78542 0.00000000 0.14545455 0.00000000 <NA>
#> 2 M70T527_POS 70.36113 526.76657 0.02564103 0.18181818 0.30000000 <NA>
#> 3 M71T775_POS 70.98125 775.44867 0.00000000 0.00000000 0.00000000 <NA>
#> 4 M71T669_POS 70.98125 668.52844 0.00000000 0.02727273 0.01818182 <NA>
#> 5 M71T715_POS 70.98125 714.74066 0.05128205 0.12727273 0.02727273 <NA>
#> 6 M71T54_POS 71.04999 54.45641 0.15384615 0.99090909 0.05454545 <NA>
#> CAS.ID HMDB.ID KEGG.ID Lab.ID Adduct mz.error mz.match.score RT.error
#> 1 <NA> <NA> <NA> <NA> <NA> NA NA NA
#> 2 <NA> <NA> <NA> <NA> <NA> NA NA NA
#> 3 <NA> <NA> <NA> <NA> <NA> NA NA NA
#> 4 <NA> <NA> <NA> <NA> <NA> NA NA NA
#> 5 <NA> <NA> <NA> <NA> <NA> NA NA NA
#> 6 <NA> <NA> <NA> <NA> <NA> NA NA NA
#> RT.match.score CE SS Total.score Database Level
#> 1 NA <NA> NA NA <NA> NA
#> 2 NA <NA> NA NA <NA> NA
#> 3 NA <NA> NA NA <NA> NA
#> 4 NA <NA> NA NA <NA> NA
#> 5 NA <NA> NA NA <NA> NA
#> 6 NA <NA> NA NA <NA> NA
table(variable_info_pos$Level)
#>
#> 1 2
#> 23 114
table(variable_info_pos$Database)
#>
#> MoNA_0.0.1 MS_0.0.2 NIST_0.0.1
#> 43 23 71
Use the ms2_plot_mass_dataset() function to get the MS2 matching plot.
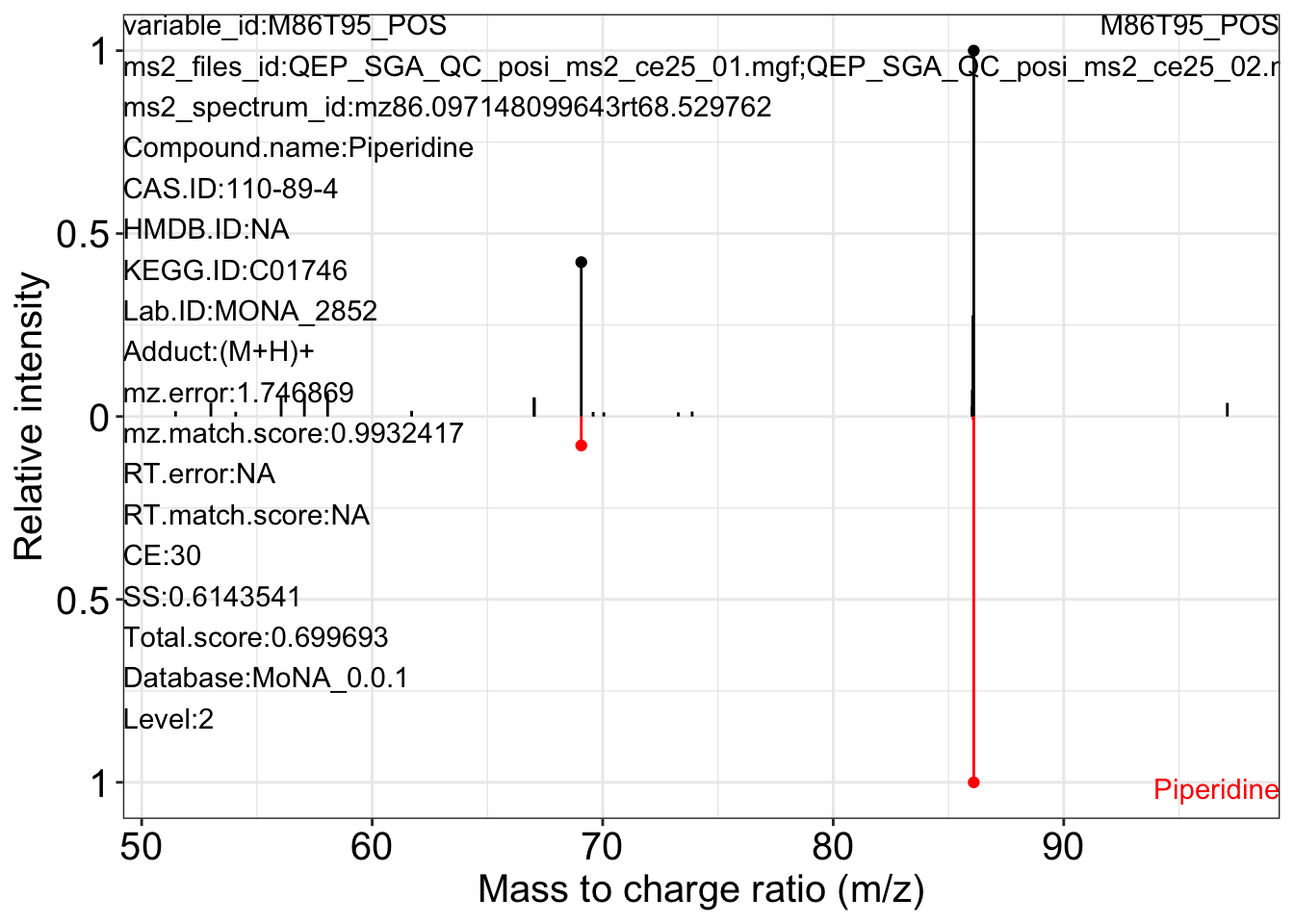
ms2_plot_mass_dataset(object = object_pos2,
variable_id = "M86T95_POS",
database = mona_database0.0.3)
#> $M86T95_POS_1
ms2_plot_mass_dataset(object = object_pos2,
variable_id = "M86T95_POS",
database = mona_database0.0.3,
interactive_plot = TRUE)
#> $M86T95_POS_1
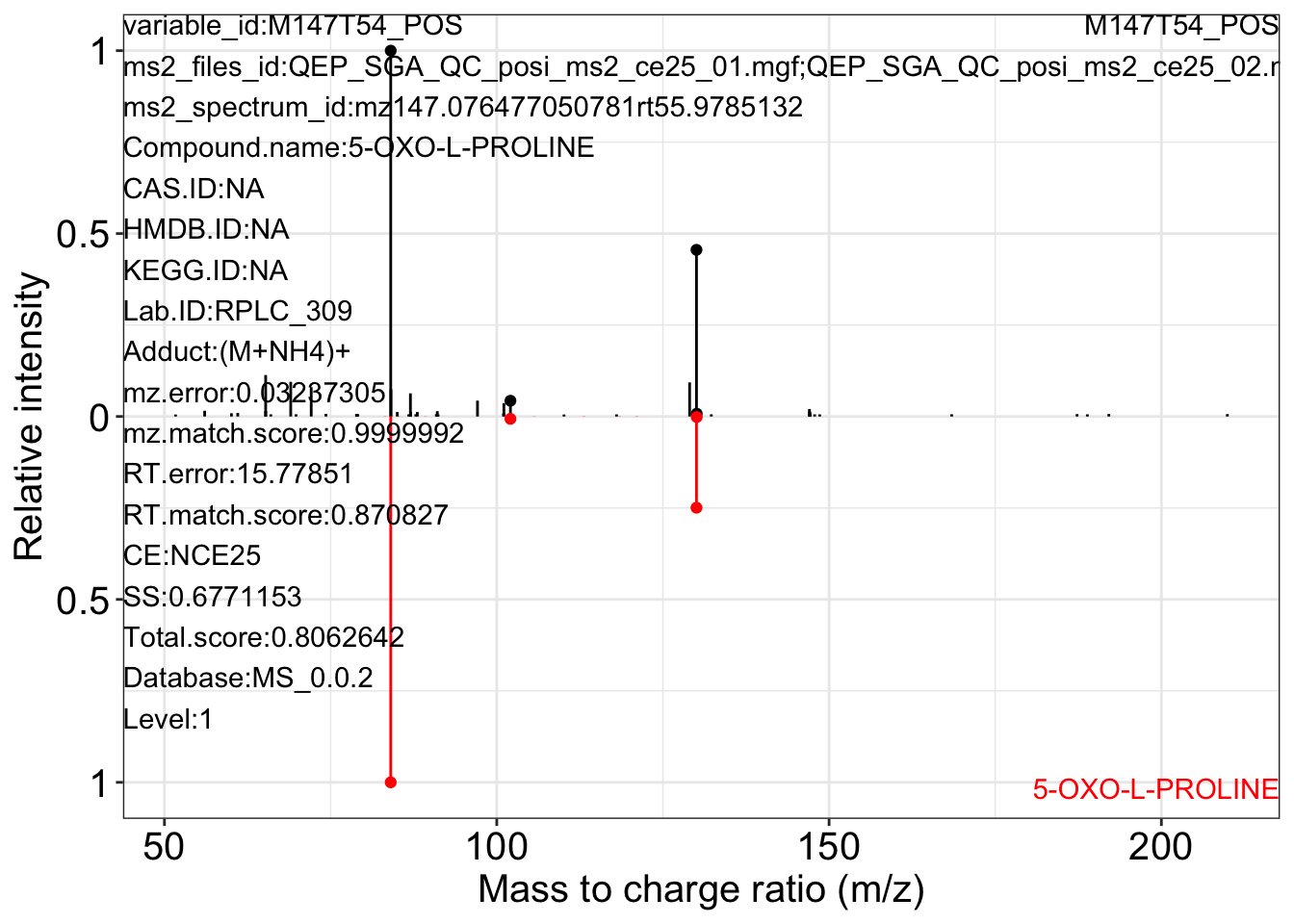
ms2_plot_mass_dataset(object = object_pos2,
variable_id = "M147T54_POS",
database = snyder_database_rplc0.0.3,
interactive_plot = FALSE)
#> database may be wrong.
#> database may be wrong.
#> $M147T54_POS_1
#>
#> $M147T54_POS_2
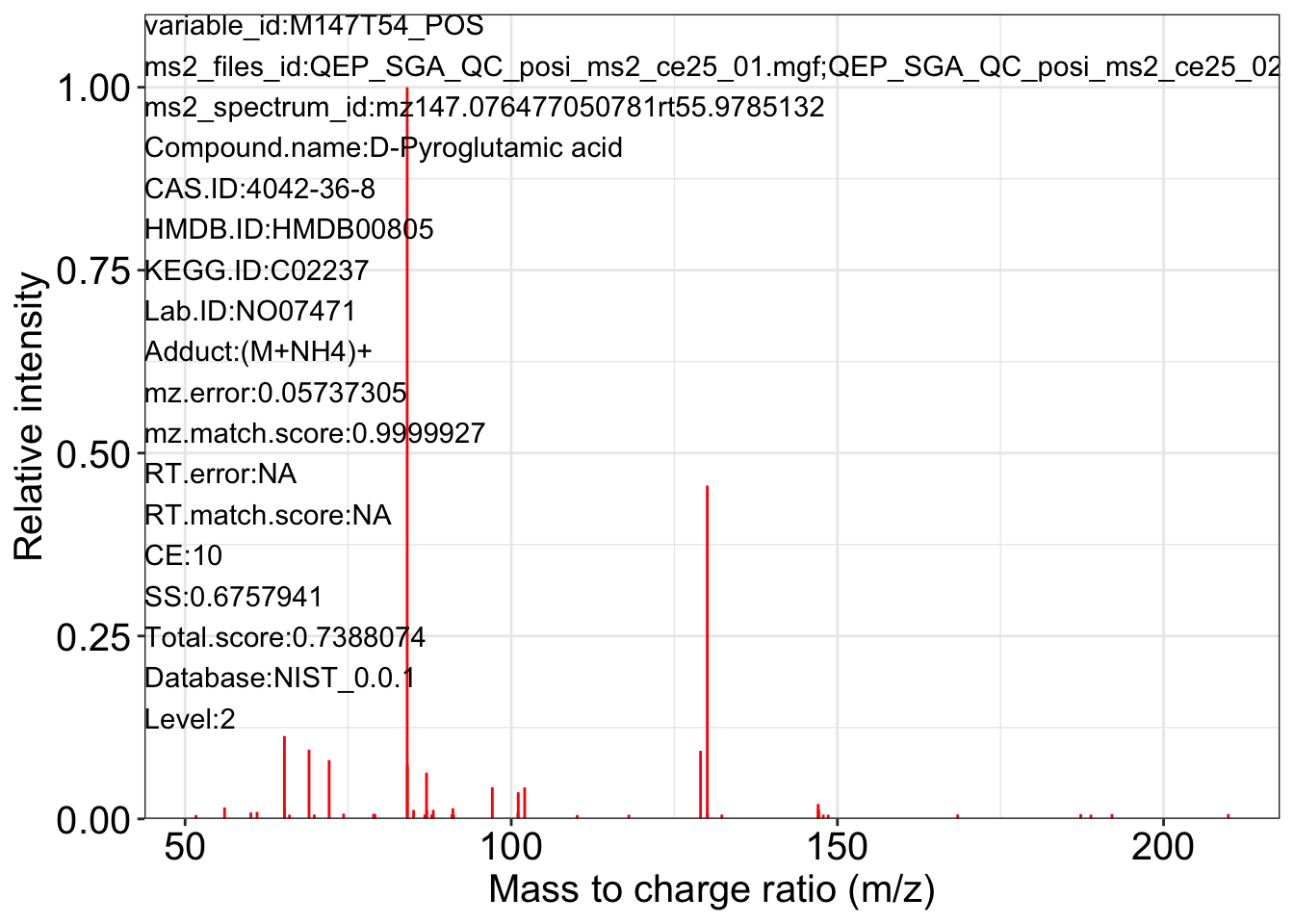
#>
#> $M147T54_POS_3
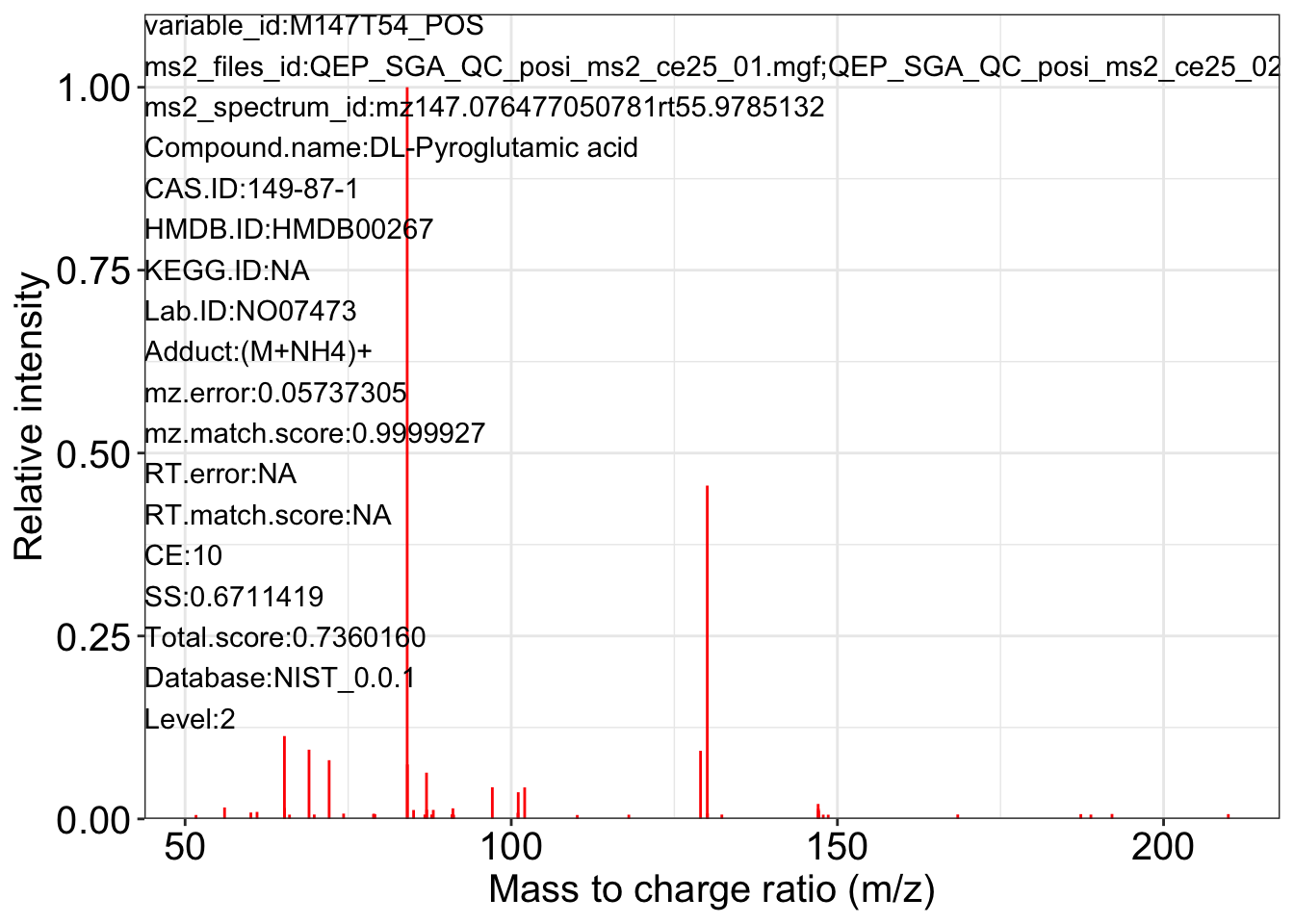
Save data for subsequent analysis.
save(object_pos2, file = "metabolite_annotation/object_pos2")
save(object_neg2, file = "metabolite_annotation/object_neg2")
Session information
sessionInfo()
#> R version 4.4.1 (2024-06-14)
#> Platform: aarch64-apple-darwin20
#> Running under: macOS 15.0
#>
#> Matrix products: default
#> BLAS: /Library/Frameworks/R.framework/Versions/4.4-arm64/Resources/lib/libRblas.0.dylib
#> LAPACK: /Library/Frameworks/R.framework/Versions/4.4-arm64/Resources/lib/libRlapack.dylib; LAPACK version 3.12.0
#>
#> locale:
#> [1] en_US.UTF-8/en_US.UTF-8/en_US.UTF-8/C/en_US.UTF-8/en_US.UTF-8
#>
#> time zone: Asia/Singapore
#> tzcode source: internal
#>
#> attached base packages:
#> [1] grid stats4 stats graphics grDevices utils datasets
#> [8] methods base
#>
#> other attached packages:
#> [1] metid_1.2.34 metpath_1.0.8 ComplexHeatmap_2.20.0
#> [4] mixOmics_6.28.0 lattice_0.22-6 MASS_7.3-61
#> [7] massstat_1.0.6 tidyr_1.3.1 ggfortify_0.4.17
#> [10] massqc_1.0.7 masscleaner_1.0.12 MSnbase_2.30.1
#> [13] ProtGenerics_1.36.0 S4Vectors_0.42.1 Biobase_2.64.0
#> [16] BiocGenerics_0.50.0 mzR_2.38.0 Rcpp_1.0.13
#> [19] xcms_4.2.3 BiocParallel_1.38.0 massprocesser_1.0.10
#> [22] ggplot2_3.5.1 dplyr_1.1.4 magrittr_2.0.3
#> [25] masstools_1.0.13 massdataset_1.0.34 tidymass_1.0.9
#>
#> loaded via a namespace (and not attached):
#> [1] fs_1.6.4 matrixStats_1.3.0
#> [3] bitops_1.0-8 fit.models_0.64
#> [5] httr_1.4.7 RColorBrewer_1.1-3
#> [7] doParallel_1.0.17 tools_4.4.1
#> [9] doRNG_1.8.6 backports_1.5.0
#> [11] utf8_1.2.4 R6_2.5.1
#> [13] lazyeval_0.2.2 GetoptLong_1.0.5
#> [15] withr_3.0.1 prettyunits_1.2.0
#> [17] gridExtra_2.3 preprocessCore_1.66.0
#> [19] cli_3.6.3 fastDummies_1.7.4
#> [21] labeling_0.4.3 sass_0.4.9
#> [23] mvtnorm_1.3-1 robustbase_0.99-4
#> [25] readr_2.1.5 randomForest_4.7-1.1
#> [27] proxy_0.4-27 pbapply_1.7-2
#> [29] foreign_0.8-87 rrcov_1.7-6
#> [31] MetaboCoreUtils_1.12.0 parallelly_1.38.0
#> [33] itertools_0.1-3 limma_3.60.4
#> [35] readxl_1.4.3 rstudioapi_0.16.0
#> [37] impute_1.78.0 generics_0.1.3
#> [39] shape_1.4.6.1 crosstalk_1.2.1
#> [41] zip_2.3.1 Matrix_1.7-0
#> [43] MALDIquant_1.22.3 fansi_1.0.6
#> [45] abind_1.4-5 lifecycle_1.0.4
#> [47] yaml_2.3.10 SummarizedExperiment_1.34.0
#> [49] SparseArray_1.4.8 crayon_1.5.3
#> [51] PSMatch_1.8.0 KEGGREST_1.44.1
#> [53] pillar_1.9.0 knitr_1.48
#> [55] GenomicRanges_1.56.1 rjson_0.2.22
#> [57] corpcor_1.6.10 codetools_0.2-20
#> [59] glue_1.7.0 pcaMethods_1.96.0
#> [61] data.table_1.16.0 remotes_2.5.0
#> [63] MultiAssayExperiment_1.30.3 vctrs_0.6.5
#> [65] png_0.1-8 cellranger_1.1.0
#> [67] gtable_0.3.5 cachem_1.1.0
#> [69] xfun_0.47 openxlsx_4.2.7
#> [71] S4Arrays_1.4.1 tidygraph_1.3.1
#> [73] pcaPP_2.0-5 ncdf4_1.23
#> [75] iterators_1.0.14 statmod_1.5.0
#> [77] robust_0.7-5 progress_1.2.3
#> [79] GenomeInfoDb_1.40.1 rprojroot_2.0.4
#> [81] bslib_0.8.0 affyio_1.74.0
#> [83] rpart_4.1.23 colorspace_2.1-1
#> [85] DBI_1.2.3 Hmisc_5.1-3
#> [87] nnet_7.3-19 tidyselect_1.2.1
#> [89] compiler_4.4.1 MassSpecWavelet_1.70.0
#> [91] htmlTable_2.4.3 DelayedArray_0.30.1
#> [93] plotly_4.10.4 bookdown_0.40
#> [95] checkmate_2.3.2 scales_1.3.0
#> [97] DEoptimR_1.1-3 affy_1.82.0
#> [99] stringr_1.5.1 digest_0.6.37
#> [101] rmarkdown_2.28 XVector_0.44.0
#> [103] htmltools_0.5.8.1 pkgconfig_2.0.3
#> [105] base64enc_0.1-3 MatrixGenerics_1.16.0
#> [107] highr_0.11 fastmap_1.2.0
#> [109] rlang_1.1.4 GlobalOptions_0.1.2
#> [111] htmlwidgets_1.6.4 UCSC.utils_1.0.0
#> [113] farver_2.1.2 jquerylib_0.1.4
#> [115] jsonlite_1.8.8 MsExperiment_1.6.0
#> [117] mzID_1.42.0 RCurl_1.98-1.16
#> [119] Formula_1.2-5 GenomeInfoDbData_1.2.12
#> [121] patchwork_1.2.0 munsell_0.5.1
#> [123] viridis_0.6.5 MsCoreUtils_1.16.1
#> [125] vsn_3.72.0 furrr_0.3.1
#> [127] stringi_1.8.4 ggraph_2.2.1
#> [129] zlibbioc_1.50.0 plyr_1.8.9
#> [131] parallel_4.4.1 listenv_0.9.1
#> [133] ggrepel_0.9.5 Biostrings_2.72.1
#> [135] MsFeatures_1.12.0 graphlayouts_1.1.1
#> [137] hms_1.1.3 Spectra_1.14.1
#> [139] circlize_0.4.16 igraph_2.0.3
#> [141] QFeatures_1.14.2 rngtools_1.5.2
#> [143] reshape2_1.4.4 XML_3.99-0.17
#> [145] evaluate_0.24.0 blogdown_1.19
#> [147] BiocManager_1.30.25 tzdb_0.4.0
#> [149] foreach_1.5.2 missForest_1.5
#> [151] tweenr_2.0.3 purrr_1.0.2
#> [153] polyclip_1.10-7 future_1.34.0
#> [155] clue_0.3-65 ggforce_0.4.2
#> [157] AnnotationFilter_1.28.0 e1071_1.7-14
#> [159] RSpectra_0.16-2 ggcorrplot_0.1.4.1
#> [161] viridisLite_0.4.2 class_7.3-22
#> [163] rARPACK_0.11-0 tibble_3.2.1
#> [165] memoise_2.0.1 ellipse_0.5.0
#> [167] IRanges_2.38.1 cluster_2.1.6
#> [169] globals_0.16.3 here_1.0.1